贝叶斯回归 - 推断算法(第 2 部分)¶
在第一部分中,我们研究了如何使用 SVI 在一个简单的贝叶斯线性回归模型上执行推断。在本教程中,我们将探讨更具表现力的指南以及精确推断技术。我们将使用与之前相同的数据集。
[1]:
%reset -sf
[2]:
import logging
import os
import torch
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from torch.distributions import constraints
import pyro
import pyro.distributions as dist
import pyro.optim as optim
pyro.set_rng_seed(1)
assert pyro.__version__.startswith('1.9.1')
[3]:
%matplotlib inline
plt.style.use('default')
logging.basicConfig(format='%(message)s', level=logging.INFO)
smoke_test = ('CI' in os.environ)
pyro.set_rng_seed(1)
DATA_URL = "https://d2hg8soec8ck9v.cloudfront.net/datasets/rugged_data.csv"
rugged_data = pd.read_csv(DATA_URL, encoding="ISO-8859-1")
贝叶斯线性回归¶
我们的目标仍然是预测一个国家的人均对数 GDP,函数关系基于数据集中的两个特征——该国家是否位于非洲,以及其地形崎岖指数,但我们将探讨更具表现力的指南。
模型 + 指南¶
我们将再次写出模型,类似于第一部分中的模型,但明确不使用 PyroModule。我们将写出回归中的每个项,使用相同的先验。bA 和 bR 是分别对应于 is_cont_africa 和 ruggedness 的回归系数,a 是截距,bAR 是两个特征之间的相关因子。
编写指南将与构建模型紧密类比,主要区别在于指南参数需要是可训练的。为此,我们使用 pyro.param() 在 ParamStore 中注册指南参数。请注意尺度参数上的正约束。
[4]:
def model(is_cont_africa, ruggedness, log_gdp):
a = pyro.sample("a", dist.Normal(0., 10.))
b_a = pyro.sample("bA", dist.Normal(0., 1.))
b_r = pyro.sample("bR", dist.Normal(0., 1.))
b_ar = pyro.sample("bAR", dist.Normal(0., 1.))
sigma = pyro.sample("sigma", dist.Uniform(0., 10.))
mean = a + b_a * is_cont_africa + b_r * ruggedness + b_ar * is_cont_africa * ruggedness
with pyro.plate("data", len(ruggedness)):
pyro.sample("obs", dist.Normal(mean, sigma), obs=log_gdp)
def guide(is_cont_africa, ruggedness, log_gdp):
a_loc = pyro.param('a_loc', torch.tensor(0.))
a_scale = pyro.param('a_scale', torch.tensor(1.),
constraint=constraints.positive)
sigma_loc = pyro.param('sigma_loc', torch.tensor(1.),
constraint=constraints.positive)
weights_loc = pyro.param('weights_loc', torch.randn(3))
weights_scale = pyro.param('weights_scale', torch.ones(3),
constraint=constraints.positive)
a = pyro.sample("a", dist.Normal(a_loc, a_scale))
b_a = pyro.sample("bA", dist.Normal(weights_loc[0], weights_scale[0]))
b_r = pyro.sample("bR", dist.Normal(weights_loc[1], weights_scale[1]))
b_ar = pyro.sample("bAR", dist.Normal(weights_loc[2], weights_scale[2]))
sigma = pyro.sample("sigma", dist.Normal(sigma_loc, torch.tensor(0.05)))
mean = a + b_a * is_cont_africa + b_r * ruggedness + b_ar * is_cont_africa * ruggedness
[5]:
# Utility function to print latent sites' quantile information.
def summary(samples):
site_stats = {}
for site_name, values in samples.items():
marginal_site = pd.DataFrame(values)
describe = marginal_site.describe(percentiles=[.05, 0.25, 0.5, 0.75, 0.95]).transpose()
site_stats[site_name] = describe[["mean", "std", "5%", "25%", "50%", "75%", "95%"]]
return site_stats
# Prepare training data
df = rugged_data[["cont_africa", "rugged", "rgdppc_2000"]]
df = df[np.isfinite(df.rgdppc_2000)]
df["rgdppc_2000"] = np.log(df["rgdppc_2000"])
train = torch.tensor(df.values, dtype=torch.float)
SVI¶
和之前一样,我们将使用 SVI 执行推断。
[6]:
from pyro.infer import SVI, Trace_ELBO
svi = SVI(model,
guide,
optim.Adam({"lr": .05}),
loss=Trace_ELBO())
is_cont_africa, ruggedness, log_gdp = train[:, 0], train[:, 1], train[:, 2]
pyro.clear_param_store()
num_iters = 5000 if not smoke_test else 2
for i in range(num_iters):
elbo = svi.step(is_cont_africa, ruggedness, log_gdp)
if i % 500 == 0:
logging.info("Elbo loss: {}".format(elbo))
Elbo loss: 5795.467590510845
Elbo loss: 415.8169444799423
Elbo loss: 250.71916329860687
Elbo loss: 247.19457268714905
Elbo loss: 249.2004036307335
Elbo loss: 250.96484470367432
Elbo loss: 249.35092514753342
Elbo loss: 248.7831552028656
Elbo loss: 248.62140649557114
Elbo loss: 250.4274433851242
[7]:
from pyro.infer import Predictive
num_samples = 1000
predictive = Predictive(model, guide=guide, num_samples=num_samples)
svi_samples = {k: v.reshape(num_samples).detach().cpu().numpy()
for k, v in predictive(log_gdp, is_cont_africa, ruggedness).items()
if k != "obs"}
让我们观察模型中不同潜在变量的后验分布。
[8]:
for site, values in summary(svi_samples).items():
print("Site: {}".format(site))
print(values, "\n")
Site: a
mean std 5% 25% 50% 75% 95%
0 9.177024 0.059607 9.07811 9.140463 9.178211 9.217098 9.27152
Site: bA
mean std 5% 25% 50% 75% 95%
0 -1.890622 0.122805 -2.08849 -1.979107 -1.887476 -1.803683 -1.700853
Site: bR
mean std 5% 25% 50% 75% 95%
0 -0.157847 0.039538 -0.22324 -0.183673 -0.157873 -0.133102 -0.091713
Site: bAR
mean std 5% 25% 50% 75% 95%
0 0.304515 0.067683 0.194583 0.259464 0.304907 0.348932 0.415128
Site: sigma
mean std 5% 25% 50% 75% 95%
0 0.902898 0.047971 0.824166 0.870317 0.901981 0.935171 0.981577
HMC¶
与使用变分推断得到潜在变量的近似后验不同,我们还可以使用马尔可夫链蒙特卡罗(MCMC)进行精确推断。MCMC 是一类算法,在极限情况下,允许我们从真实的后验中抽取无偏样本。我们将使用的算法是 No-U Turn Sampler (NUTS) [1],它提供了一种高效且自动运行 Hamiltonian Monte Carlo 的方法。它比变分推断稍慢,但提供了精确的估计。
[9]:
from pyro.infer import MCMC, NUTS
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_samples=1000, warmup_steps=200)
mcmc.run(is_cont_africa, ruggedness, log_gdp)
hmc_samples = {k: v.detach().cpu().numpy() for k, v in mcmc.get_samples().items()}
Sample: 100%|██████████| 1200/1200 [00:30, 38.99it/s, step size=2.76e-01, acc. prob=0.934]
[10]:
for site, values in summary(hmc_samples).items():
print("Site: {}".format(site))
print(values, "\n")
Site: a
mean std 5% 25% 50% 75% 95%
0 9.182098 0.13545 8.958712 9.095588 9.181347 9.277673 9.402615
Site: bA
mean std 5% 25% 50% 75% 95%
0 -1.847651 0.217768 -2.19934 -1.988024 -1.846978 -1.70495 -1.481822
Site: bR
mean std 5% 25% 50% 75% 95%
0 -0.183031 0.078067 -0.311403 -0.237077 -0.185945 -0.131043 -0.051233
Site: bAR
mean std 5% 25% 50% 75% 95%
0 0.348332 0.127478 0.131907 0.266548 0.34641 0.427984 0.560221
Site: sigma
mean std 5% 25% 50% 75% 95%
0 0.952041 0.052024 0.869388 0.914335 0.949961 0.986266 1.038723
比较后验分布¶
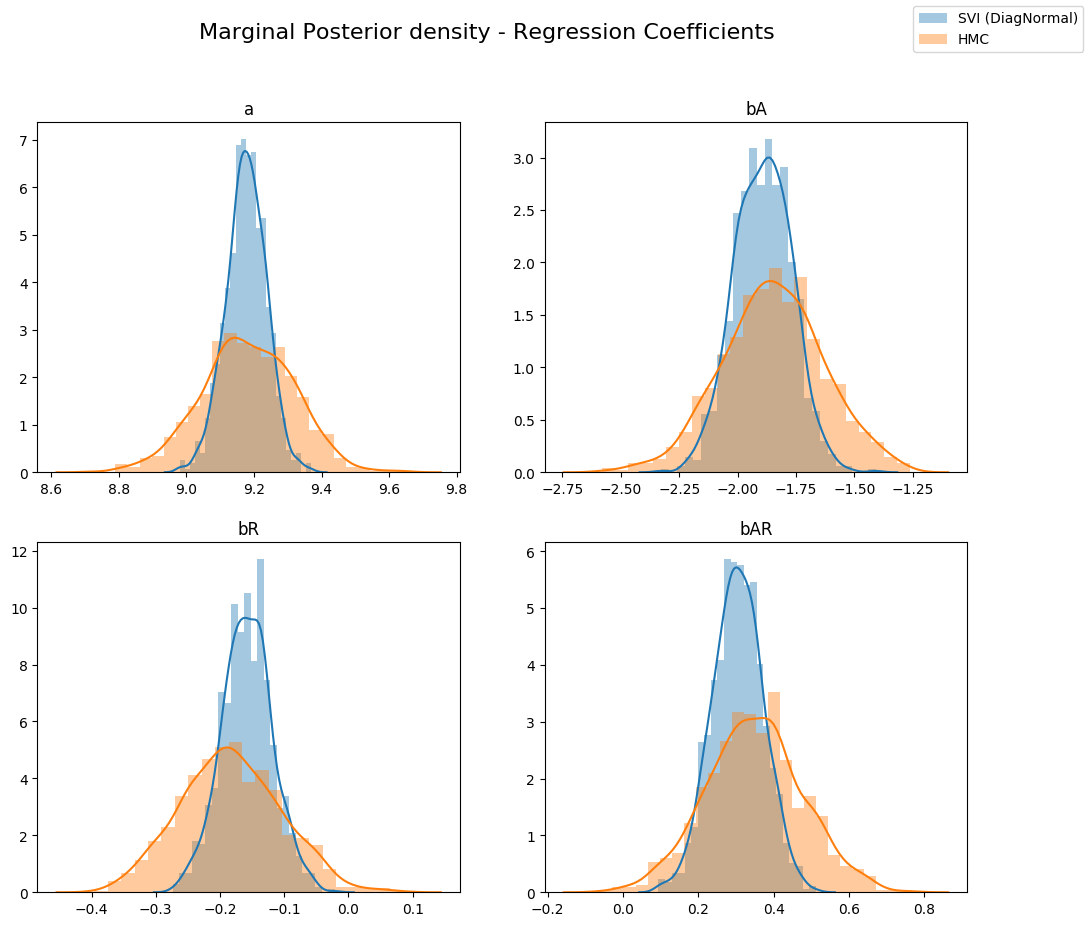
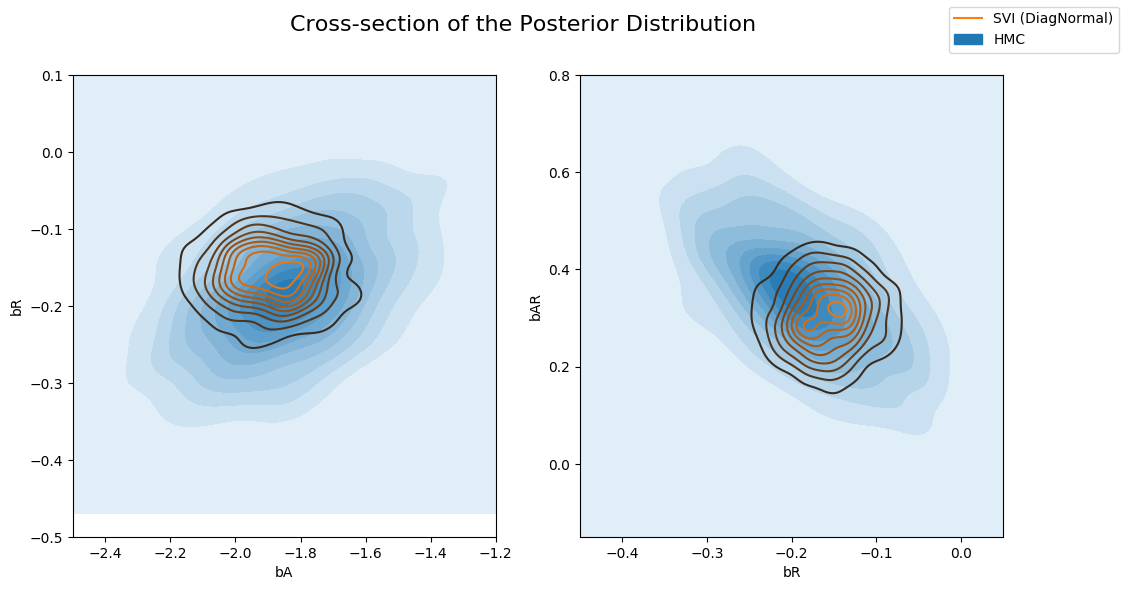
让我们比较从变分推断获得的潜在变量的后验分布与从 Hamiltonian Monte Carlo 获得的后验分布。如下所示,对于变分推断,不同回归系数的边缘分布相对于真实后验(来自 HMC)是过度集中的(under-dispersed)。这是变分推断最小化的 KL(q||p) 损失(真实后验与近似后验之间的 KL 散度)的一个副作用。
当我们将联合后验分布的不同横截面与变分推断的近似后验叠加绘制时,这一点可以更清楚地看出。请注意,由于我们的变分族具有对角协方差,我们无法建模潜在变量之间的任何相关性,结果得到的近似是过度自信的(过度集中)。
[11]:
sites = ["a", "bA", "bR", "bAR", "sigma"]
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(12, 10))
fig.suptitle("Marginal Posterior density - Regression Coefficients", fontsize=16)
for i, ax in enumerate(axs.reshape(-1)):
site = sites[i]
sns.distplot(svi_samples[site], ax=ax, label="SVI (DiagNormal)")
sns.distplot(hmc_samples[site], ax=ax, label="HMC")
ax.set_title(site)
handles, labels = ax.get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right');
[12]:
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))
fig.suptitle("Cross-section of the Posterior Distribution", fontsize=16)
sns.kdeplot(x=hmc_samples["bA"], y=hmc_samples["bR"], ax=axs[0], shade=True, label="HMC")
sns.kdeplot(x=svi_samples["bA"], y=svi_samples["bR"], ax=axs[0], label="SVI (DiagNormal)")
axs[0].set(xlabel="bA", ylabel="bR", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))
sns.kdeplot(x=hmc_samples["bR"], y=hmc_samples["bAR"], ax=axs[1], shade=True, label="HMC")
sns.kdeplot(x=svi_samples["bR"], y=svi_samples["bAR"], ax=axs[1], label="SVI (DiagNormal)")
axs[1].set(xlabel="bR", ylabel="bAR", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))
handles, labels = axs[1].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right');
MultivariateNormal 指南¶
作为与之前从 Diagonal Normal 指南获得结果的比较,我们现在将使用一个从多元正态分布的 Cholesky 分解中生成样本的指南。这允许我们通过协方差矩阵捕获潜在变量之间的相关性。如果手动编写,我们需要组合所有潜在变量,以便联合采样多元正态。
[13]:
from pyro.infer.autoguide import AutoMultivariateNormal, init_to_mean
guide = AutoMultivariateNormal(model, init_loc_fn=init_to_mean)
svi = SVI(model,
guide,
optim.Adam({"lr": .01}),
loss=Trace_ELBO())
is_cont_africa, ruggedness, log_gdp = train[:, 0], train[:, 1], train[:, 2]
pyro.clear_param_store()
for i in range(num_iters):
elbo = svi.step(is_cont_africa, ruggedness, log_gdp)
if i % 500 == 0:
logging.info("Elbo loss: {}".format(elbo))
Elbo loss: 703.0100790262222
Elbo loss: 444.6930855512619
Elbo loss: 258.20718491077423
Elbo loss: 249.05364602804184
Elbo loss: 247.2170884013176
Elbo loss: 247.28261297941208
Elbo loss: 246.61236548423767
Elbo loss: 249.86004841327667
Elbo loss: 249.1157277226448
Elbo loss: 249.86634194850922
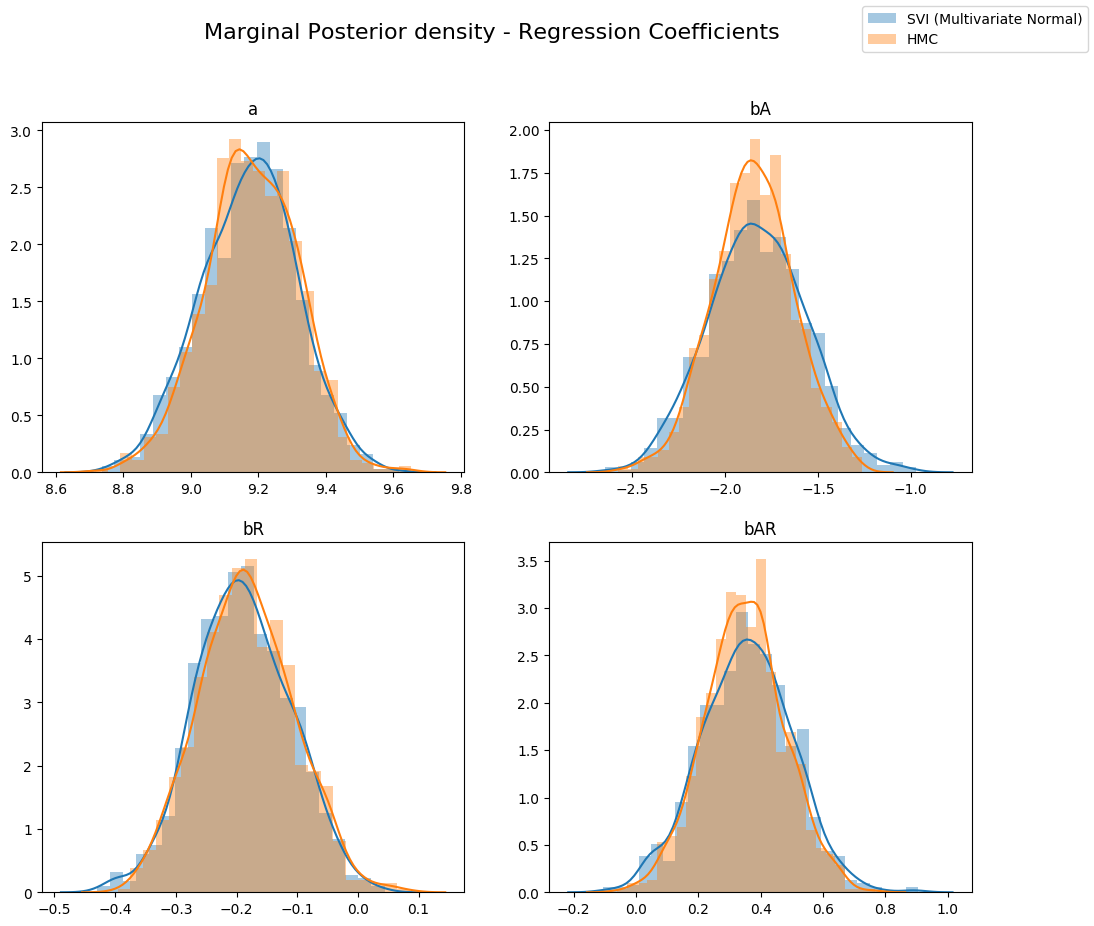
让我们再次查看后验的形状。您可以看到多元指南能够捕获真实后验的更多信息。
[14]:
predictive = Predictive(model, guide=guide, num_samples=num_samples)
svi_mvn_samples = {k: v.reshape(num_samples).detach().cpu().numpy()
for k, v in predictive(log_gdp, is_cont_africa, ruggedness).items()
if k != "obs"}
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(12, 10))
fig.suptitle("Marginal Posterior density - Regression Coefficients", fontsize=16)
for i, ax in enumerate(axs.reshape(-1)):
site = sites[i]
sns.distplot(svi_mvn_samples[site], ax=ax, label="SVI (Multivariate Normal)")
sns.distplot(hmc_samples[site], ax=ax, label="HMC")
ax.set_title(site)
handles, labels = ax.get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right');
现在让我们比较 Diagonal Normal 指南与 Multivariate Normal 指南计算的后验。请注意,多元分布比 Diagonal Normal 分布更分散。
[15]:
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))
fig.suptitle("Cross-sections of the Posterior Distribution", fontsize=16)
sns.kdeplot(x=svi_samples["bA"], y=svi_samples["bR"], ax=axs[0], label="SVI (Diagonal Normal)")
sns.kdeplot(x=svi_mvn_samples["bA"], y=svi_mvn_samples["bR"], ax=axs[0], shade=True, label="SVI (Multivariate Normal)")
axs[0].set(xlabel="bA", ylabel="bR", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))
sns.kdeplot(x=svi_samples["bR"], y=svi_samples["bAR"], ax=axs[1], label="SVI (Diagonal Normal)")
sns.kdeplot(x=svi_mvn_samples["bR"], y=svi_mvn_samples["bAR"], ax=axs[1], shade=True, label="SVI (Multivariate Normal)")
axs[1].set(xlabel="bR", ylabel="bAR", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))
handles, labels = axs[1].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right');
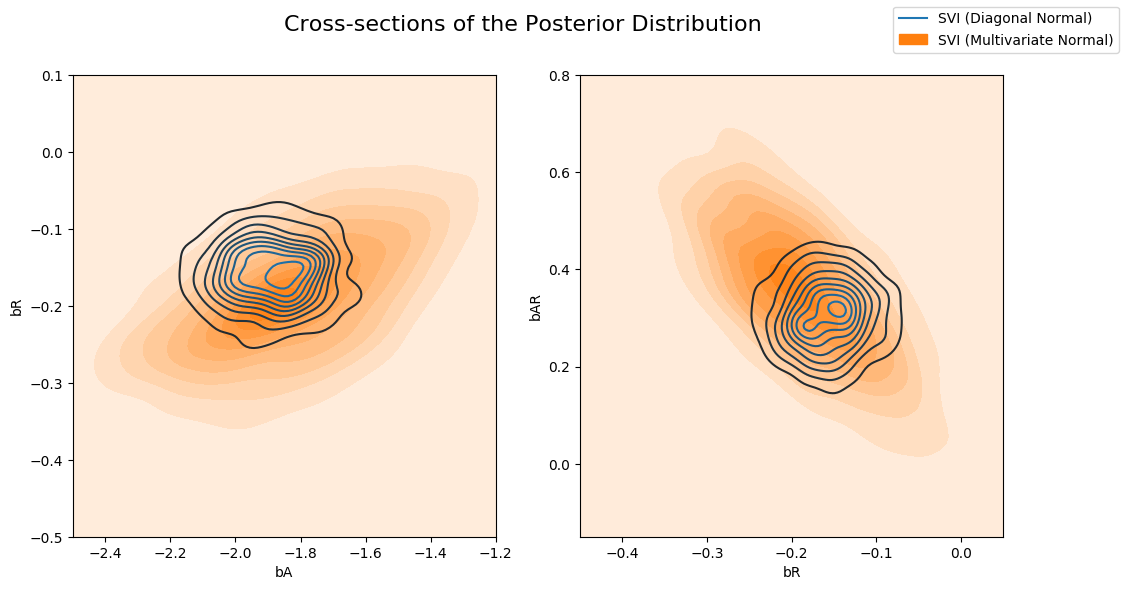
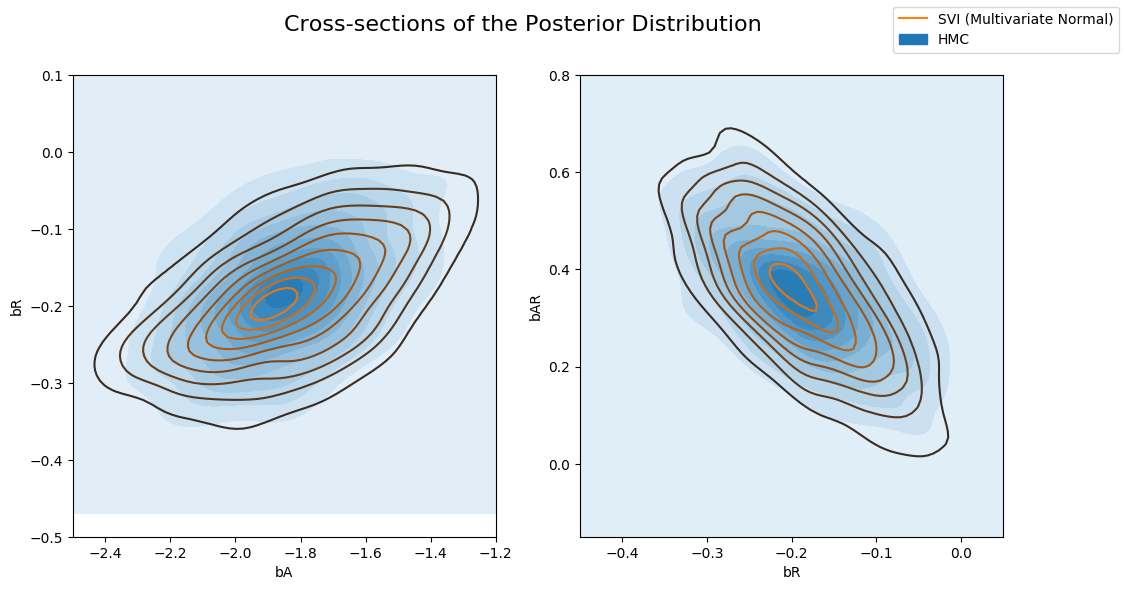
以及 Multivariate 指南与 HMC 计算的后验。请注意,Multivariate 指南更好地捕获了真实后验。
[16]:
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))
fig.suptitle("Cross-sections of the Posterior Distribution", fontsize=16)
sns.kdeplot(x=hmc_samples["bA"], y=hmc_samples["bR"], ax=axs[0], shade=True, label="HMC")
sns.kdeplot(x=svi_mvn_samples["bA"], y=svi_mvn_samples["bR"], ax=axs[0], label="SVI (Multivariate Normal)")
axs[0].set(xlabel="bA", ylabel="bR", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))
sns.kdeplot(x=hmc_samples["bR"], y=hmc_samples["bAR"], ax=axs[1], shade=True, label="HMC")
sns.kdeplot(x=svi_mvn_samples["bR"], y=svi_mvn_samples["bAR"], ax=axs[1], label="SVI (Multivariate Normal)")
axs[1].set(xlabel="bR", ylabel="bAR", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))
handles, labels = axs[1].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right');
参考文献¶
[1] Hoffman, Matthew D., and Andrew Gelman. “The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo.” Journal of Machine Learning Research 15.1 (2014): 1593-1623. https://arxiv.org/abs/1111.4246。